Paper category: Original research paper
Corresponding author: Marcin Kuciński (marcin_kucinski1@o2.pl)
DOI: 10.2478/oandhs-2021-0005
Received: 16/06/2020
Accepted: 31/08/2020
Full text: here
Citation (APA style): Kuciński, M. & Fopp-Bayat, D. (2021). Phylogenetic characteristics of selected European huchen (Hucho hucho L.) broodstocks – implication for broodstock management. Oceanological and Hydrobiological Studies, 50(1), 38-46. https://doi.org/10.2478/oandhs-2021-0005
Abstract
European huchen (Hucho hucho) is a representative of large and rare migratory salmonid fish, which has become endangered due to extensive anthropogenic changes in freshwater ecosystems. Numerous broodstocks of the European huchen have therefore been established throughout the species’ range in recent years to supplement wild fisheries of this species. Unfortunately, this conservation management strategy entails a number of potential ecological and genetic risks associated with the release of farm-raised fish into wild populations.
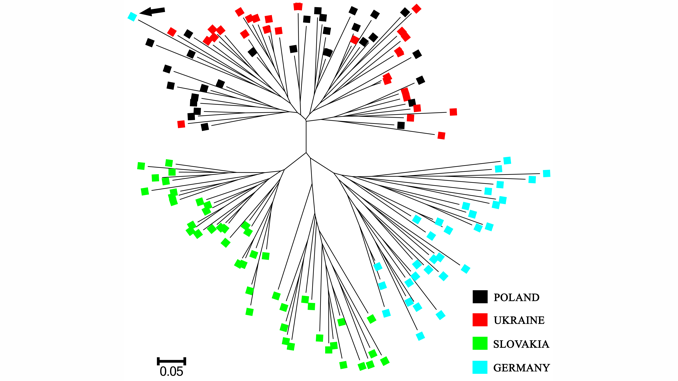
A comprehensive and feasible genetic monitoring protocol for broodstocks maintained for the production of restocking material is therefore essential in the sustainable management of critically endangered fish species. The current paper presents phylogenetic characteristics of four selected huchen broodstocks across Central and Eastern Europe. Genetic comparisons of the studied broodstocks were based on ten microsatellite DNA markers. The effective population size (Ne), the individual assignment test, the Principal Coordinates Analysis (PCoA), the allele sharing distance (DAS) and the Bayesian clustering analysis were applied in this study. Moreover, five selected fragments of mitochondrial DNA were used for molecular verification of species membership and genetic purity
of examined specimens.
Acknowledgements
The authors thank the anonymous reviewers for their comments and suggestions on the manuscript. This study was supported by projects No. GW/2013/12 (Optimization of PCR parameters of selected mitochondrial DNA fragments) and 2014/15/N/NZ9/01515 (Amplification of microsatellite DNA fragments and genotyping) funded by the University of Warmia and Mazury in Olsztyn and the National Research Centre in Poland.
We would like to sincerely thank Prof. Kolman R., Dr. Ocalewicz K., Dr. Svinger V.W., Dr. Lebeda I. and Liszewski T. for their help in sampling and genotypic data analysis.
References
Araki, H., Waples, R.S., Ardren, W.R. & Cooper, B. (2007). Effective population size of steelhead trout: influence of variance in reproductive success, hatchery programs, and genetic compensation between life-history forms. Mol. Ecol. 16(5): 953–966. DOI: 10.1111/j.1365-294X.2006.03206.x.
Bernas, R., Burzynski, A., Debowski, P., Pocwierz-Kotus, A. & Wenne, R. (2014). Genetic diversity within sea trout population from an intensively stocked southern Baltic river, based on microsatellite DNA analysis. Fisheries Manag. Ecol. 21: 398–409. DOI: 10.1111/fme.12090.
Bowcock, A.M., Ruiz-Linares, A., Tomfohrde, J., Minch, E., Kidd, J.R. et al. (1994). High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368(6470): 455–457. DOI: 10.1038/368455a0.
Ditlecadet, D., Dufresne, F., Le Francois, N.R. & Blier, P.U. (2006). Applying microsatellites in two commercial strains of Arctic charr (Salvelinus alpinus): Potential for selective breeding program. Aquaculture 257(1): 37-43. DOI: 10.1016/j.aquaculture.2006.03.016.
Do, C., Waples, R.S., Peel, D., Macbeth, G.M., Tillet, B.J. et al. (2013). NeEstimator V2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Res. 14(1): 209–214. DOI: 10.1111/1755-0998.12157.
Drywa, A., Pocwierz-Kotus, A., Was, A., Dobosz, S., Kent, M.P. et al. (2013). Genetyping of two populations of Southern Baltic Sea trout Salmo trutta m. trutta using an Atlantic salmon derived SNP-array. Mar. Genomics. 9: 25–32. DOI: 10.1016/j.margen.2012.08.001.
Einum, S. & Fleming, I.A. (2001). Implication of stocking: ecological interaction between wild related salmonids. Nor. J. Fresh. Res. 75: 56–70.
Evanno, G., Regnaut, S. & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620. DOI: 10.1111/j.1365-294X.2005.02553.x.
Falush, D., Stephens, M. & Pritchard, J.K. (2003). Inference of population structure using multilocus genotypic data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
FishBase 2015. Retrieved May 15, 2020 from http://www.fishbase.org.
Fopp-Bayat, D., Jankun, M., Kuźmiński, H. (2010). Genetic characterization of Polish cultured brook trout, Salvelinus fontinalis (Mitchill), based on microsatellite DNA analysis. Arch. Pol. Fisheries. 18: 93–99. DOI: 10.2478/v10086-010-0011-2.
Frankham, R., Ballou, J.D., David, A. & Briscoe D.A. (2010). Introduction to Conservation Genetics, 2nd ed. Cambridge University Press, Cambridge.
Geist, J., Kolasha, M., Gum, B. & Kuehn, R. (2009). The importance of genetic cluster recognition for the conservation of migratory fish species: the example of the endangered European huchen Hucho hucho (L.). J. Fish. Biol. 75: 1063–1078. DOI: 10.1111/j.1095-8649.2009.02377.x.
Geist, J. (2011). Integrative freshwater ecology and biodiversity conservation. Ecol. Indic. 11: 1507–1516. DOI: 10.1016/j.ecolind.2011.04.002.
Glover, K.A., Taggart, J.B., Skaala, O. & Teale, A.J. (2004). A study of inadvertent domestication selection during start-feeding of brown trout families. J. Fish. Biol. 64: 1168–1178. DOI: 10.1111/j.0022-1112.2004.00376.x.
Hansen, M.M. (2002). Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Mol. Ecol. 11: 1003–1015. DOI: 10.1046/j.1365-294X.2002.01495.x.
Hoarou, G., Boon, E., Jongma, D.N., Ferber, S., Palsson, J., van der Veer, H.W. et al. (2005). Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L.). P. Roy. Soc. B-Biol. Sci. 272: 497–503. DOI: 10.1098/rspb.2004.2963.
Huff, D.D., Miller, L.M., Chizinski, C.J. & Vondracek, B. (2011). Mixed-source reintroductions lead to outbreeding depression in second generation descendants of a native North American fish. Mol. Ecol. 20(20): 4246–4258. DOI: 10.1111/j.1365-294X.2011.05271.x.
IUCN. (2015). Red list of Threatened Species. Retrieved June 15, 2015 from http://www.iucnredlist.org.
Kaczmarczyk, D., Luczyński, M. & Brzuzan, P. (2012). Genetic variation in three paddlefish (Polyodon spathula Walbaum) stocks based on microsatellite DNA analysis. Czech J. Anim. Sci. 57(8): 345–352.
Kalinowski, S.T., Manlove, K.R. & Taper, M.L. (2007). ONCOR: A Computer Program for Genetic Stock Identification. Bozeman, MT: Department of Ecology. Montana State University. Retrived June, 15, 2015 from http://www.montana.edu/kalinowski/Software/ONCOR.htm.
Kottelat, M. & Freyhof, J. (2007). Handbook of European freshwater fishes. Switzerland: Steven Simpson Books.
Kucinski, M., Fopp-Bayat, D., Liszewski, T., Svinger, V. Lebeda, I. et al. (2015a). Genetic analysis of four European huchen (Hucho hucho Linnaeus, 1758) broodstocks from Poland, Germany, Slovakia and Ukraine: implication for conservation. J. App. Genet. 56(4): 469–480. DOI: 10.1007/s13353-015-0274-9.
Kucinski, M., Fopp-Bayat, D., Zivna, D., Liszewski, T., Svinger, V. et al. (2015b). Application of mtDNA markers for European huchen (Hucho hucho Linnaeus, 1758) management in Poland. Czech J.. Anim. Sci. 60: 564–569. DOI: 10.17221/8599-CJAS.
Langella, O. (2002). Populations 1.2.28. Logiciel de génétique des populations. Laboratoire Populations, génétique et évolution, CNRS UPR 9034, Gif-sur-Yvette. Retrived May 15, 2020 from http://bioinformatics.org/~tryphon/populations/#ancre_bibliographie.
Maric, S., Razpet, A., Nikolic, V. & Simonovic, P. (2011). Genetic differentiation of European grayling (Thymallus thymallus) populations in Serbia, based on mitochondrial and nuclear DNA analyses. Genet. Sel. Evol. 42: 2. DOI: 10.1186/1297-9686-43-2.
Nei, M., Tajima, F. & Tateno, Y. (1983). Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 19: 153–170. DOI: 10.1007/BF02300753.
Peakall, R. & Smouse, P.E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. DOI: 10.1093/bioinformatics/bts460.
Piry, S., Alapetite, A., Cornuet, J.-M., Paetkau, D., Baudouin, L. & Estoup, A. (2004). GeneClass2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 95: 536–539. DOI: 10.1093/jhered/esh074.
Pocwierz-Kotus, A., Bernas, R., Kent, M.P., Lien, S., Leliuna, E. et al. (2015). Restitution and genetic differentiation of salmon populations in the southern Baltic genetyped with the Atlantic salmon 7K SNP array. Genet. Sel. Evol. 47: 39. DOI: 10.1186/s12711-015-0121-9.
Pritchard, J.K., Stephens, M. & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Slatkin, M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462.
Sonstebo, J.H. Borgstrom, R. & Heun, M. (2007). Genetic structure of brown trout (Salmo trutta L.) from the Hardangervidda mountain plateau (Norway) analyzed by microsatellite DNA: a basis for conservation guideline. Conserv. Genet. 8: 33–44. DOI: 10.1007/s10592-006-9145-6.
Tringali, M.D. & Bert, T.M. (1998). Risk to genetic effective population size should be an important consideration in fish stock-enhancement programs. Bull. Mar. Sci. 62(2): 641–659.
Vaha, J.P., Erkinaro, J., Niemela, E. & Primmer, C.R. (2007). Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol. Ecol. 16: 2638–2658. DOI: 10.1111/j.1365-294X.2007.03329.x.
Van Oosterhout, C., Hutchinson, W.F., Wills, D.P.M. & Shipley, P. (2004). Micro-Checker: software for identifying and correcting genotypes errors in microsatellite data. Mol. Ecol. Notes. 4(3): 535–538. DOI: 10.1111/j.1471-8286.2004.00684.x.
Vrijenhoek, R.C. (1998). Conservation genetics of freshwater fish. J. Fish. Biol. 53: 394–412. DOI: 10.1111/j.1095-8649.1998.tb01039.x.
Walsh, P.S., Metzger, D.A. & Higuchi, R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10: 506–513.
Wang, S., Hard, J.J. & Utter, F. (2002). Salmonid inbreeding: a review. Rev. Fish. Biol. Fisher. 11: 301–319. DOI: 10.1023/A:1021330500365.
Waples, R.S. (1999). Dispelling some myths about hatcheries. Fisheries 24: 12–21. DOI: 10.1577/1548-8446(1999)0242.0.CO;2.
Wedekind, C., Rudolfsen, G., Jacob, A., Urbach, D. & Muller, R. (2007). The genetic consequences of hatchery-induced competition in a salmonid. Biol. Conserv. 137: 180–188. DOI: 10.1016/j.biocon.2007.01.025.
Weir, B.S. & Cockerman, C.C. (1984). Estimating F statistics for the analysis of population structure. Evolution 38: 1358–1370. DOI: 10.2307/2408641.
Wenburg, J.K., Bentzen, P. & Foote, C.J. (1998). Microsatellite analysis of genetic population structure in an endangered salmonid: the coastal cutthroat trout (Oncorhynchus clarki). Mol. Ecol. 7: 733–749. DOI: 10.1046/j.1365-294x.1998.00386.x.
Yang, X., Qian, L., Wu, H., Fan, Z. & Wang, C. (2012). Population differentiation, bottleneck and selection of Eurasian perch (Perca fluviatilis L.) at the Asian edge of its natural range. Biochem. Syst. Ecol. 40: 6–12. DOI: 10.1016/j.bse.2011.09.002.