Paper category: Original research paper
Corresponding author: Zhao-Yu Jiang (jiangzhaoyu@lyu.edu.cn)
DOI: 10.1515/ohs-2020-0003
Received: 25/04/2019
Accepted: 26/08/2019
Full text: here
Citation (APA style): Jiang, Z. & Sun, F. (2020). Diversity and biogeography of picoplankton communities from the Straits of Malacca to the South China Sea. Oceanological and Hydrobiological Studies, 49(1), pp. 23-33. Retrieved 10 Mar. 2020, from doi:10.1515/ohs-2020-0003
Abstract
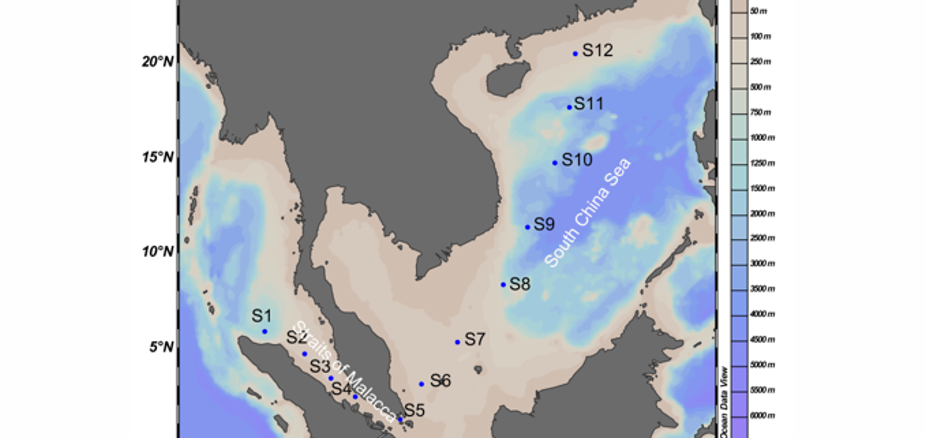
Marine picoplankton, including prokaryotic and eukaryotic picoplankton, drive many biogeochemical processes, such as carbon, nitrogen and sulfur cycles, making them crucial to the marine ecosystem. Despite the fact that picoplankton is prevalent, its diversity and spatial distribution from the Straits of Malacca (SM) to the South China Sea (SCS) remain poorly investigated. This work explores the phylogenetic diversity and community structure of picoplankton in relation to environmental factors from the SM to the SCS. To this end, the Illumina MiSeq sequencing technique was applied to 16S and 18S rRNA genes. The results showed significant differences in the dynamics of picoplankton between the open sea and the strait region. Proteobacteria and Cyanobacteria constituted a larger part of the prokaryotic group. Within Cyanobacteria, the abundance of Prochlorococcus in the open sea was significantly higher than that of Synechococcus, while the opposite trend was observed in the strait. Dinoflagellata, Cnidaria, Retaria, Tunicata, and Arthropoda dominated among the eukaryotic taxa. High-throughput sequencing data indicated that salinity, temperature and NO<sup>2</sup>-N were the key factors determining the prokaryotic community structure, while temperature and dissolved oxygen determined the eukaryotic community structure in the studied region. The network analysis demonstrated that the cooperation and competition were also important factors affecting the picoplankton community.
References
Amaral-Zettler, L.A., McCliment, E.A., Ducklow, H.W. & Huse, S.M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PloS one 4(7): e6372. DOI: 10.1371/journal.pone.0006372.
Annett, A.L., Carson, D.S., Crosta, X., Clarke, A. & Ganeshram, R.S. (2010). Seasonal progression of diatom assemblages in surface waters of Ryder Bay, Antarctica. Polar Biology 33(1): 13–29. DOI: 10.1007/s00300-009-0681-7.
Buitenhuis, E.T., Li, W.K., Vaulot, D., Lomas, M.W., Landry, M.R. (2012). Picophytoplankton biomass distribution in the global ocean. Earth System Science Data 4(1): 37–46. DOI: 10.5194/essd-4-37-2012.
Campbell, B.J., Yu, L., Heidelberg, J.F., & Kirchman, D.L. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proceedings of the National Academy of Sciences 108(31): 12776–12781. DOI: 10.1073/pnas.1101405108.
Caporaso, J.G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F.D. et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7(5): 335–336. DOI: 10.1038/nmeth.f.303.
Cerino, F., Aubry, F.B., Coppola, J., La Ferla, R., Maimone, G. et al. (2012). Spatial and temporal variability of pico-, nano- and microphytoplankton in the offshore waters of the southern Adriatic Sea (Mediterranean Sea). Continental Shelf Research 44: 94–105. DOI: 10.1016/j.csr.2011.06.006.
Chen, B., Liu, H., Huang, B. & Wang, J. (2014). Temperature effects on the growth rate of marine picoplankton. Marine Ecology Progress Series 505: 37–47. DOI: 10.3354/meps10773.
Chen, B., Wang, L., Song, S., Huang, B., Sun, J. et al. (2011). Comparisons of picophytoplankton abundance, size, and fluorescence between summer and winter in northern South China Sea. Continental Shelf Research 31(14): 1527–1540. DOI: 10.1016/j.csr.2011.06.018.
Chen, C.C., Shiah, F.K., Chung, S.W., & Liu, K.K. (2006). Winter phytoplankton blooms in the shallow mixed layer of the South China Sea enhanced by upwelling. Journal of Marine Systems 59(1–2): 97–110. DOI: 10.1016/j.jmarsys.2005.09.002.
Cram, J.A., Chow, C.E.T., Sachdeva, R., Needham, D.M., Parada, A.E. et al. (2015). Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. The ISME Journal 9(3): 563–580. DOI: 10.1038/ismej.2014.153.
De Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F. et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348(6237): 1261605. DOI: 10.1126/science.1261605.
del Campo, J., Guillou, L., Hehenberger, E., Logares, R., López-García, P. et al. (2016). Ecological and evolutionary significance of novel protist lineages. European Journal of Protistology 55: 4–11. DOI: 10.1016/j.ejop.2016.02.002.
Edgar, R.C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19): 2460–2461. DOI: 10.1093/bioinformatics/btq461.
Flombaum, P., Gallegos, J.L., Gordillo, R.A., Rincón, J., Zabala, L.L. et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proceedings of the National Academy of Sciences 110(24): 9824–9829. DOI: 10.1073/pnas.1307701110.
Fuchs, B.M., Woebken, D., Zubkov, M.V., Burkill, P., & Amann, R. (2005). Molecular identification of picoplankton populationsin contrasting waters of the Arabian Sea. Aquatic Microbial Ecology 39(2): 145–157. DOI: 10.3354/ame039145.
Halsey, K.H., Giovannoni, S.J., Graus, M., Zhao, Y., Landry, Z. et al. (2017). Biological cycling of volatile organic carbon by phytoplankton and bacterioplankton. Limnology and Oceanography 62(6): 2650–2661. DOI: 10.1002/lno.10596.
Huang, D., Licuanan, W.Y., Hoeksema, B.W., Chen, C.A., Ang, P.O. et al. (2015). Extraordinary diversity of reef corals in the South China Sea. Marine Biodiversity 45(2): 157–168. DOI: 10.1007/s12526-014-0236-1.
Jiang, Z.Y., Wang, Y.S., Cheng, H., Sun, C.C., & Wu, M.L. (2015). Variation of phytoplankton community structure from the Pearl River estuary to South China Sea. Ecotoxicology 24(7–8): 1442–1449. DOI: 10.1007/s10646-015-1494-9.
Jiang, Z.Y., Wang, Y.S., & Sun, F.L. (2014). Spatial structure of eukaryotic ultraplankton community in the northern South China Sea. Biologia 69(5): 557–565. DOI: 10.2478/s11756-014-0361-0.
Kayal, E., Roure, B., Philippe, H., Collins, A.G. & Lavrov, D.V. (2013). Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evolutionary Biology 13(1): 5. DOI: 10.1186/1471-2148-13-5.
Li, J., Jiang, X., Li, G., Jing, Z., Zhou, L. et al. (2017). Distribution of picoplankton in the northeastern South China Sea with special reference to the effects of the Kuroshio intrusion and the associated mesoscale eddies. Science of the Total Environment 589: 1–10. DOI: 10.1016/j.scitotenv.2017.02.208.
Liang, Y., Zhang, Y., Zhang, Y., Luo, T., Rivkin, R.B. et al. (2017). Distributions and relationships of virio-and picoplankton in the epi-, meso- and bathypelagic zones of the Western Pacific Ocean. FEMS Microbiology Ecology 93(2): fiw238. DOI: 10.1093/femsec/fiw238.
Lin, L., Wang, Y.S., Sun, C.C., Li, N., Wang, H. et al. (2011). Demonstration of a new indicator for studying upwelling in the northern South China Sea. Oceanologia 53(2): 605–622. DOI: 10.5697/oc.53-2.605.
Ling, J., Dong, J.D., Wang, Y.S., Zhang, Y.Y., Deng, C. et al. (2012). Spatial variation of bacterial community structure of the Northern South China Sea in relation to water chemistry. Ecotoxicology 21(6): 1669–1679. DOI: 10.1007/s10646-012-0941-0.
Mackinson, B.L., Moran, S.B., Lomas, M.W., Stewart, G.M., & Kelly, R.P. (2015). Estimates of micro-, nano-, and picoplankton contributions to particle export in the northeast Pacific. Biogeosciences 12(11): 3429–3446. DOI: 10.5194/bg-12-3429-2015.
Magoč, T. & Salzberg, S.L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21): 2957–2963. DOI: 10.1093/bioinformatics/btr507.
Man-Aharonovich, D., Philosof, A., Kirkup, B.C., Le Gall, F., Yogev, T. et al. (2010). Diversity of active marine picoeukaryotes in the Eastern Mediterranean Sea unveiled using photosystem-II psbA transcripts. The ISME Journal 4(8): 1044–1052. DOI: 10.1038/ismej.2010.25.
Meador, T.B., Gogou, A., Spyres, G., Herndl, G.J., Krasakopoulou, E. et al. (2010). Biogeochemical relationships between ultrafiltered dissolved organic matter and picoplankton activity in the Eastern Mediterranean Sea. Deep Sea Research Part II: Topical Studies in Oceanography 57(16): 1460–1477. DOI: 10.1016/j.dsr2.2010.02.015.
Mena, C., Reglero, P., Ferriol, P., Torres, A.P., Aparicio-González, A. et al. (2016). Prokaryotic picoplankton spatial distribution during summer in a haline front in the Balearic Sea, Western Mediterranean. Hydrobiologia 779(1): 243–257. DOI: 10.1007/s10750-016-2825-4.
Montoya, J.M., Pimm, S.L., & Solé, R.V. (2006). Ecological networks and their fragility. Nature 442(7100): 259–264. DOI: 10.1038/nature04927.
Moon-van der Staay, S.Y., De Wachter, R. & Vaulot, D. (2001). Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409(6820): 607–610. DOI: 10.1038/35054541.
Morton, B. & Blackmore, G. (2001). South China Sea. Marine Pollution Bulletin 42(12): 1236–1263. DOI: 10.1016/S0025-326X(01)00240-5.
Ning, X.R., Chai, F., Xue, H., Cai, Y., Liu, C. et al. (2004). Physical‐biological oceanographic coupling influencing phytoplankton and primary production in the South China Sea. Journal of Geophysical Research: Oceans 109(C10): C1005. DOI: 10.1029/2004JC002365.
Price, D.C. & Bhattacharya, D. (2017). Robust Dinoflagellata phylogeny inferred from public transcriptome databases. Journal of Phycology 53(3): 725–729. DOI: 10.1111/jpy.12529.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K.S. et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(7285): 59–65. DOI: 10.1038/nature08821.
Richardson, T.L., & Jackson, G.A. (2007). Small phytoplankton and carbon export from the surface ocean. Science 315(5813): 838–840. DOI: 10.1126/science.1133471.
Rii, Y.M., Duhamel, S., Bidigare, R.R., Karl, D.M., Repeta, D.J. et al. (2016). Diversity and productivity of photosynthetic picoeukaryotes in biogeochemically distinct regions of the South East Pacific Ocean. Limnology and Oceanography 61(3): 806–824. DOI: 10.1002/lno.10255.
Rocha, J., Calado, R. & Leal, M. (2015). Marine Bioactive Compounds from Cnidarians. In: S.K. Kim (Ed.) Springer Handbook of Marine Biotechnology (pp. 823–849). Springer, Berlin, Heidelberg.
Schattenhofer, M., Fuchs, B.M., Amann, R., Zubkov, M.V., Tarran, G.A. et al. (2009). Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environmental Microbiology 11(8): 2078–2093. DOI: 10.1111/j.1462-2920.2009.01929.x.
Schattenhofer, M., Wulf, J., Kostadinov, I., Glöckner, F.O., Zubkov, M.V. et al. (2011). Phylogenetic characterisation of picoplanktonic populations with high and low nucleic acid content in the North Atlantic Ocean. Systematic and Applied Microbiology 34(6): 470–475. DOI: 10.1016/j.syapm.2011.01.008.
Schloss, P.D., Westcott, S.L., Ryabin, T., Hall, J.R., Hartmann, M. et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology 75(23): 7537–7541. DOI: 10.1128/AEM.01541-09.
Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T. et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13(11): 2498–2504. DOI: 10.1101/gr.1239303.
Shi, R., Li, J., Qi, Z., Zhang, Z., Liu, H. et al. (2018) Abundance and community composition of bacterioplankton in the Northern South China Sea during winter: geographic position and water layer influences. Biologia 73(2): 197–206. DOI: 10.2478/s11756-018-0023-8.
Suh, S.S., Park, M., Hwang, J., Lee, S., Chung, Y. et al. (2014). Distinct patterns of marine bacterial communities in the South and North Pacific Oceans. Journal of Microbiology 52(10): 834–841. DOI: 10.1007/s12275-014-4287-6.
Sun, F.L., Wang, Y.S., Wu, M.L., Sun, C.C. & Cheng, H. (2015). Spatial and vertical distribution of bacterial community in the northern South China Sea. Ecotoxicology 24(7–8): 1478–1485. DOI: 10.1007/s10646-015-1472-2.
Sunagawa, S., Coelho, L.P., Chaffron, S., Kultima, J.R., Labadie, K. et al. (2015). Structure and function of the global ocean microbiome. Science 348(6237): 1261359. DOI: 10.1126/science.1261359.
Sze, Y., Miranda, L.N., Sin, T.M., & Huang, D. (2018). Characterising planktonic dinoflagellate diversity in Singapore using DNA metabarcoding. Metabarcoding and Metagenomics 2: e25136. DOI: 10.3897/mbmg.2.25136.
Tamm, M., Laas, P., Freiberg, R., Nõges, P. & Nõges, T. (2018). Parallel assessment of marine autotrophic picoplankton using flow cytometry and chemotaxonomy. Science of the Total Environment 625: 185–193. DOI: 10.1016/j.scitotenv.2017.12.234.
Taylor, F.J.R., Hoppenrath, M., & Saldarriaga, J.F. (2008). Dinoflagellate diversity and distribution. Biodiversity and Conservation 17(2): 407–418. DOI: 10.1007/s10531-007-9258-3.
Wang, G., Chen, D. & Su, J. (2008). Winter eddy genesis in the eastern South China Sea due to orographic wind jets. Journal of Physical Oceanography 38(3): 726–732. DOI: 10.1175/2007JPO3868.1.
Wang, Q., Garrity, G.M., Tiedje, J.M., & Cole, J.R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73(16): 5261–5267. DOI: 10.1128/AEM.00062-07.
Wu, W., Huang, B., Liao, Y. & Sun, P. (2014). Picoeukaryotic diversity and distribution in the subtropical–tropical South China Sea. FEMS Microbiology Ecology 89(3): 563–579. DOI: 10.1111/1574-6941.12357.
Xenopoulos, M.A., Downing, J.A., Kumar, M.D., Menden‐Deuer, S. & Voss, M. (2017). Headwaters to oceans: Ecological and biogeochemical contrasts across the aquatic continuum. Limnology and Oceanography 62(S1): S3–S14. DOI: 10.1002/lno.10721.
Xia, X., Guo, W. & Liu, H. (2015). Dynamics of the bacterial and archaeal communities in the Northern South China Sea revealed by 454 pyrosequencing of the 16S rRNA gene. Deep Sea Research Part II: Topical Studies in Oceanography 117: 97–107. DOI: 10.1016/j.dsr2.2015.05.016.
Xiao, W., Wang, L., Laws, E., Xie, Y., Chen, J. et al. (2018). Realized niches explain spatial gradients in seasonal abundance of phytoplankton groups in the South China Sea. Progress in Oceanography 162: 223–239. DOI: 10.1016/j.pocean.2018.03.008.
Xiong, J., Ye, X., Wang, K., Chen, H., Hu, C. et al. (2014). Biogeography of the sediment bacterial community responds to a nitrogen pollution gradient in the East China Sea. Applied and Environmental Microbiology 80(6): 1919–1925. DOI: 10.1128/AEM.03731-13.
Zhang, Y., Jiao, N. & Hong, N. (2008). Comparative study of picoplankton biomass and community structure in different provinces from subarctic to subtropical oceans. Deep Sea Research Part II: Topical Studies in Oceanography 55(14–15): 1605–1614. DOI: 10.1016/j.dsr2.2008.04.014.
Zhang, Y., Zhao, Z., Dai, M., Jiao, N. & Herndl, G.J. (2014). Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Molecular Ecology 23(9): 2260–2274. DOI: 10.1111/mec.12739.
Zhou, J., Deng, Y., Luo, F., He, Z., Tu, Q. et al. (2010). Functional molecular ecological networks. MBio 1(4): e00169-10. DOI: 10.1128/mBio.00169-10.
Zubkov, M.V., Sleigh, M.A., & Burkill, P.H. (2000). Assaying picoplankton distribution by flow cytometry of underway samples collected along a meridional transect across the Atlantic Ocean. Aquatic Microbial Ecology 21(1): 13–20. DOI: 10.3354/ame021013.
Zwirglmaier, K., Jardillier, L., Ostrowski, M., Mazard, S., Garczarek, L. et al. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environmental Microbiology 10(1): 147–161.

Bądź pierwszy, który skomentuje ten wpis